NIST23 now available!
About
- EI library adds 40K more compounds with retention indices, most selected, measured, and evaluated by NIST.
- MS/MS tandem library grown to 51K compounds, all at multiple fragmentation energies and conditions. 66% more compounds. 80% more spectra. 2.4M spectra.
- NIST MS Search 3.0 (new release) made significant enhancements to its metadata and software.
- Upgrade discount for any previous versions
- Increased coverage and quality in all libraries (EI MS, MS/MS, and GC RI). Nearly 2x increase in MS/MS (1.3M), 40K more EI, 40K more GI-RI. New EI compounds added for importance and measured at NIST, thoroughly evaluated.
- NIST MS updated: GC retention index estimated, new compound selection process, new library building method, new evaluation process, MS interpreter updates.
- Upgrade discount for any previous versions
- Increased coverage and quality in all libraries (EI MS, MS/MS, and GC RI). Nearly 3x increase in MS/MS.
- New and improved search algorithms (e.g. hybrid search)
- Please see Update Registration to receive e-mail notifications when future updates to your NIST and/or Wiley MS libraries are available.
Benefits:
- Identify unknown compounds from EI (GC/MS) and MS/MS spectra, using library searching.
- Hundreds of thousands of EI MS spectra, MS/MS spectra, GC data & retention indices, and/or chemical names/structures.
- World's most widely used and trusted mass spectral reference library.
- Search software included. Supports most MS software systems (Agilent ChemStation/MassHunter, Thermo Xcalibur/Chromeleon, Shimadzu GCMS Solutions, Waters MassLynx, Thermo Xcalibur, PE TurboMass, etc.).
Description
The NIST 23 (2023) mass spectral library, the successor to the NIST 20/17/14, is a fully evaluated collection of electron ionization (EI) and MS/MS mass spectra, with chemical and GC data, plus search software to identify your own unknown spectra. It is a product of a more than three decade, comprehensive evaluation and expansion of the world's most widely used mass spectral reference library by a team of experienced mass spectrometrists at the National Institute of Standards and Technology (NIST) in which each spectrum was examined for correctness.
NIST 23 Components
NIST 23 is not just a mass spectral library. It contains these components:
- Electron ionization (EI) mass spectral library - ~394K carefully evaluated spectra of 347,100 unique compounds, with identifications, nearly all with chemical structures. 40K increase from NIST 20 (largest EI increase ever). You may search names of NIST compounds online.
- MS/MS library - 2.4 million spectra of 51K compounds (60% more than NIST 20), 186K precursor ions. Covers a variety of fragmentation methods: 49,590 HRAM (high res accurate mass) compounds, 50,071 QTOF/HCD/IT-HRAM/QqQ compounds, 49,561 ion trap (low res, up to MS4) compounds, 561 APCOI HRAM compounds. Variety of precursor ion types: 44,191 protonated, 19,620 deprotonated, 14,318 water/ammonia loss, 44,547 other in-source generated. (Also available separately as the NIST MS/MS Database.)
- Gas Chromatography (GC) data library - 492K retention index values (Kovats/Lee), 153K compounds in the EI library, covering both polar and non-polar columns. 40K increase from NIST 17. Includes retention indices with GC column conditions and literature citations. (Also available separately as the NIST GC Database.)
- NIST MS Search software (version 3.0) - software for searching (identifying) compounds from their mass spectra and for browsing mass spectral libraries. Also includes MS interpretation programs for analyzing mass spectra on the basis of chemical structure, molecular formula, isotopic patterns, and more.
- AMDIS software - software for deconvoluting gas/liquid chromatograms
- Documentation - Approximately 50 page electronic manual on setup and basic usage. Additional information is in the help files.
Libraries are formatted the binary format suitable for use alone or by the NIST MS Search software (and AMDIS). Additional instrument-specific formats (e.g. Agilent ChemStation and MassHunter) are available separately to permit library searching directly within your GC/MS or LC/MS data system.
With each purchase, we send an electronic download (installs software, libraries, and documentation). NIST23MF is USB stick.
Online Search (NIST)
To check for compounds in the NIST library, you may search names of compounds online.
Summary Statistics
Statistics for NIST versions are given below.
| NIST 23 | NIST 20 | NIST 17 | NIST 14 | NIST 11 | NIST 08 | NIST 05 | NIST 02 | |
|---|---|---|---|---|---|---|---|---|
| EI Spectral Library: Number of EI Spectra |
394,054(*) | 350,643(*) | 306,622(*) | 276,248(*) | 243,893(*) | 220,460(*) (with separable collection of 717 spectra of 672 salts) |
190,825(*) | 174,948 |
| Number of Unique Compounds | 347,100 | 306,869 | 267,376 | 242,466 | 212,961 | 192,108 | 163,198 | 147,198 |
| Retention Index (Kovats/Lee) | 491,790 index values for 180,618 compounds (153,472 in the EI library) | 447,285 index values for 139,693 compounds (111,788 in the EI library) | 404,045 index values for 99,400 compounds (72,361 in the EI library) | 387,463 index values for 82,868 compounds (56,216 in the EI library) | 346,757 index values for 70,838 compounds (38,648 in the EI library) | 224,038 index values for 44,008 compounds (21,847 in the EI library), now on both polar and non-polar columns. | 121,112 index values for 25,893 compounds (12,452 in the EI Library), on only non-polar columns. | NONE |
| MS/MS Library |
2,374,064 spectra of 399,267 precursor ions for 51,501 compounds. 49,590 HRAM (High Res Accurate Mass) compounds, 50,071 QTOF/HCD/IT-HRAM/QqQ compounds, 49,561 Ion Trap Compounds (Low Res., up to MS4) compounds, 561 APCI HRAM compounds. Precursor Ion Types: 44,191 Protonated, 19,620 Deprotonated, 14,318 Water/Ammonia Loss, 44,547 Other In-Source Generated. |
1,320,389 spectra of 185,608 precursor ions for 30,999 compounds. Fragmentation Methods: 27,840 HRAM (High Res Accurate Mass) Compounds; 29,890 QTOF, HCD, IT-HRAM, QqQ Compounds; 29,444 Ion Trap Compounds (Low Res, up to MS4); 246 APCI HRAM 'Extractables and Leachables'. Precursor ion types: 26,575 Protonated, 12,589 Deprotonated, 10,032 Water/Ammonia Loss, 24,167 Other In-Source Generated. |
652,475 spectra: 176,594 ion trap spectra for 120,346 different ions of 14,351 compounds, and 475,881 collision cell spectra (qtof and tandem quad) for 39,158 different ions of 14,073 compounds. | 234,284 spectra: 51,216 ion trap spectra for 42,126 different ions of 8,171 compounds, and 183,068 collision cell spectra (qtof and tandem quad) spectra for 14,835 different ions of 7,692 compounds. | 95,409 spectra (10,065 ion trap spectra of 9,194 different ions of 4,628 compounds; 85,344 collision cell spectra (qtof and tandem quad) of 7,172 different ions of 3,877 compounds)[#] (121,586 spectra in 2012 update) | 14,802 spectra of 5,308 precursor ions (3,898 cations and 1,410 anions). | 5,191 spectra of 1,943 different ions (1,671 positive and 341 negative) | NONE |
| NIST MS Search software | v. 3.0 (2023-06) | v. 2.4 (2020-06) | v. 2.3 (2017-06) | v. 2.2 (2014-04) | v. 2.0g (2011-05) | v. 2.0f (2008-08) | v. 2.0d (2005-04) | v. 2.0a (2002-07) |
| AMDIS software | v. 2.73 (2017-06) | v. 2.73 (2017-06) | v. 2.73 (2017-06) | v. 2.72 (2014-04) | v. 2.70 (2011-05) | v. 2.65 (2006-12) | v. 2.62 (2005-03) | v. 2.1 (2002-07) |
| Release Date | June 2023 | June 2020 | June 2017 | July 2014 | June 2011 | August 2008 | June 2005 | July 2002 |
(*) plus quality improvements
See also What's New in NIST 23.
Electron Ionization (EI) mass spectral library
This is a "fully evaluated" collection of electron ionization (EI) mass spectra. It is the product of a more than three decade, comprehensive evaluation and expansion of the world's most widely used mass spectral reference library by a team of experienced mass spectrometrists in which each spectrum was examined for correctness. Each spectrum has been carefully evaluated, and all decisions regarding selection or deletion were made only after agreement of two experienced mass spectral evaluators, as described in presentations at major conferences. While computer methods assisted in finding chemical identification errors and inconsistencies, and revealed certain varieties of mass spectral errors, manual interpretation was the principal basis for this evaluation effort.
The NIST Electron Ionization (EI) mass spectral library consists of 394K spectra of 347K unique compounds. Besides spectra, typical data include name, formula, molecular structure (.mol), molecular weight, CAS number, contributor name, list of peaks, synonyms, and estimated and/or measured retention index.
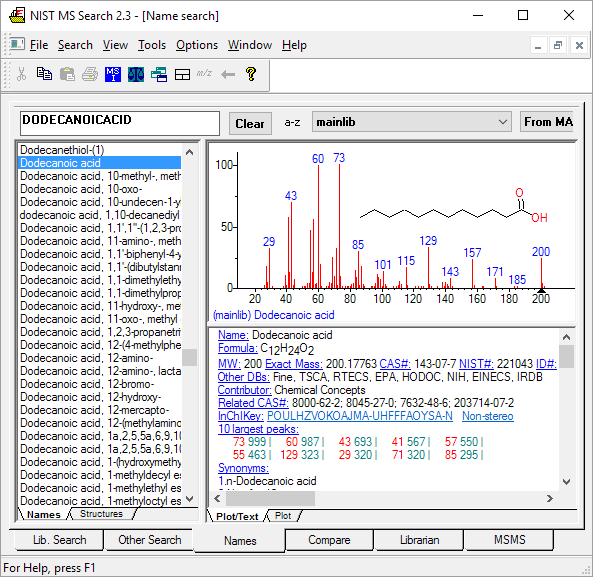
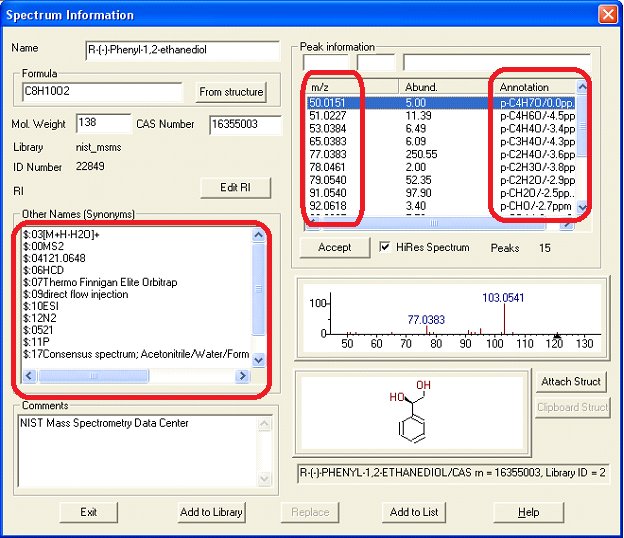
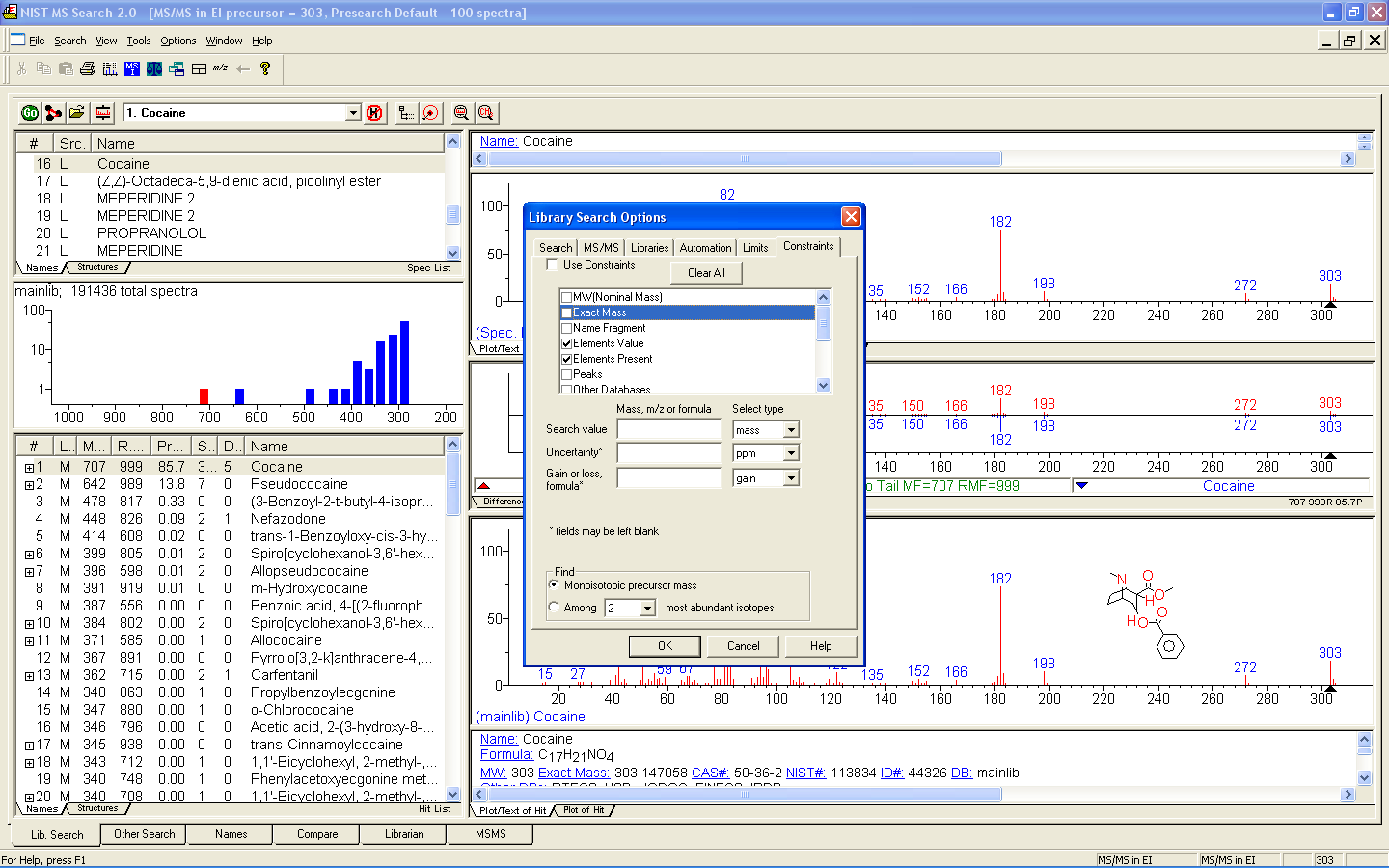
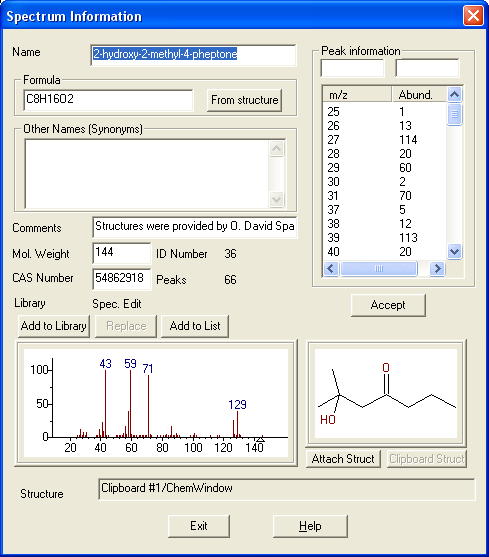
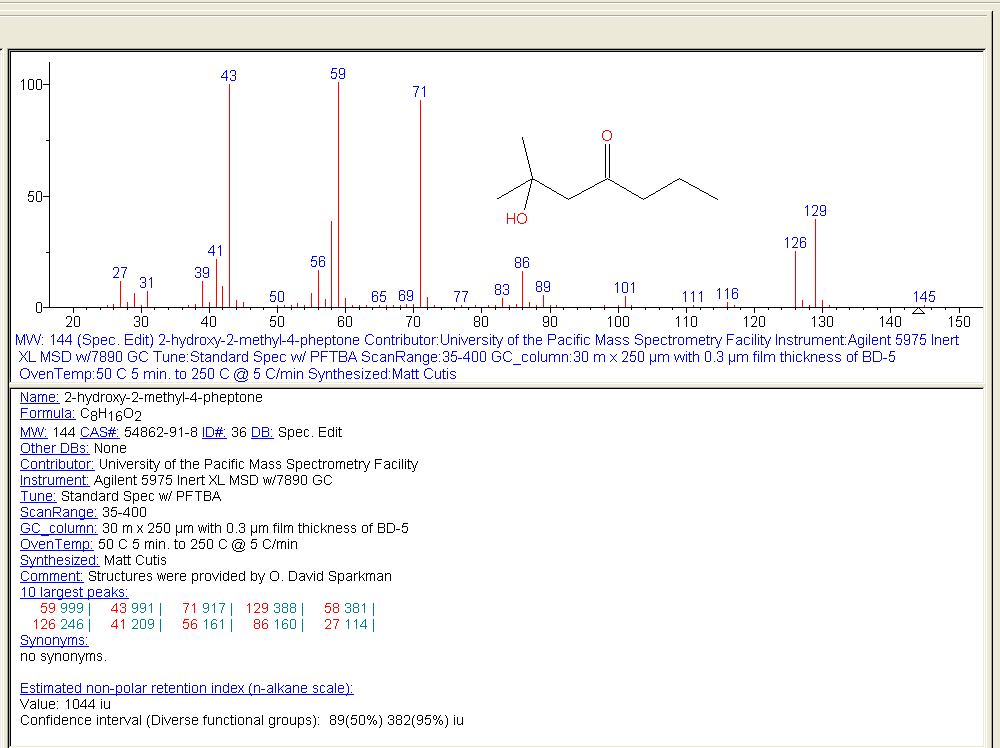
Figure: NIST MS Search software showing EI mass spectrum and compound information.
Name: 1-Decanol Formula: C10H22O MW: 158 Exact Mass: 158.167066 CAS#: 112-30-1 NIST#: 374910 ID#: 37364 DB: mainlib Other DBs: Fine, TSCA, RTECS, HODOC, NIH, EINECS, IRDB Contributor: NIST Mass Spectrometry Data Center, 2010 InChIKey: MWKFXSUHUHTGQN-UHFFFAOYSA-N 10 largest peaks: 70 999 | 55 987 | 56 859 | 69 823 | 43 732 | 41 727 | 83 724 | 84 476 | 57 452 | 97 389 | Synonyms: 1.Decyl alcohol; 2.n-Decan-1-ol; 3.n-Decanol; 4.n-Decyl alcohol; 5.Alcohol C10; 6.Alfol 10; 7.Capric alcohol; 8.Caprinic alcohol; 9.Decanol; 10.Nonylcarbinol; 11.Sipol L10; 12.T-148; 13.Decylic Alcohol; 14.Decan-1-ol; 15.Decanol-(1); 16.Agent 504; 17.Antak; 18.Dytol S-91; 19.Decyl, n- alcohol; 20.Lorol 22; 21.Primary decyl alcohol; 22.Royaltac; 23.C 10 alcohol; 24.Epal 10; 25.Royaltac-85; 26.Royaltac M-2; 27.Lorol C10; 28.Nonylcacarbinol; 29.1-Hydroxydecane; 30.Conol 10N; 31.Kalcohl 10H; 32.NSC 406313; 33.Nacol 10-99
The NIST EI Library focuses on:
- Drugs, metabolites, and poisons,
- Pesticides and fungicides,
- Organics present in soil, water and air,
- Aminoacids, di- & tri-peptides,
- Common sample contaminants,
- Common analytical derivatives of the above
Here is a breakdown by mass: (NIST 17)
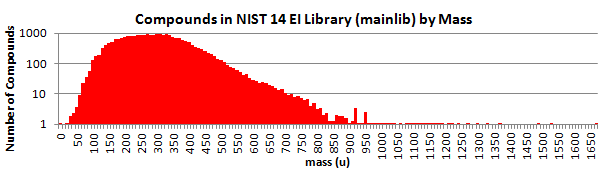
The best quality spectra are placed in the Main Library; and good-quality, alternate spectra are provided in the Selected Replicates Library.
NIST includes a collection salts and low-volatile substances not expected to be measurable by GC/MS. Here's a breakdown by mass (data from 2008 version, of 717 spectra of 672 compounds):
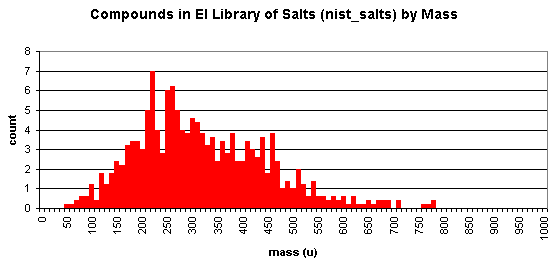
To check whether certain compounds or classes of compounds exist within the NIST database, you may do an online search of compounds in the NIST MS database.
NIST spectra have been obtained from thousands of sources. Major sources include
# Spectra, Name (data from 2014 version)
44447 V.G.Zaikin,R.S.Borisov, Topchiev Inst. Petrochem. Synth (TIPS RAS), Moscow
30498 A.A.Kutin, Moscow, Russia
29729 Chemical Concepts
22527 NIST Mass Spectrometry Data Center
31(2014), 199(2013), 1347(2012), 270(2011), 4034(2010), 1126(2009), 60(2008),
237(2007), 1335(1998), 48(1995), 2775(1994), 211(1992), 2183(1990), 8672(other)
16737 Japan AIST/NIMC Database: 9026(MS-NW-X), 7711(MS-IW-X)
11135 L.N.Kulikova,R.S.Borisov,V.G.Zaikin, Peoples' Friendship University of Russia, Moscow
9136 J. Little, Eastman chemical company,Kingsport,TN
7895 Div. of Experiment Therapeutics WRAIR, WRAMC, Washington DC 20307
4794 V.A.Korolev, Moscow, Russia
4581 A.T.Lebedev, Moscow State University, Russia
3760 LAC, NIDDK, NIH, Bethesda, MD 20892
2836 D.HENNEBERG, MAX-PLANCK INSTITUTE, MULHEIM, WEST GERMANY
2807 ASES Database, Dalian Institute, P.R. China
2735 CARL DJERASSI DEPT OF CHEM STANFORD UNIV STANFORD CALIF 94305
2308 Asinex Ltd, Moscow, Russia
1886 Institute of Organic Chemistry, USSR: 1988,1990
1862 A.Mazepa, Phys.-Chem.Institute, Odessa, Ukraine
1728 RADIAN CORP
1683 Humboldt-Universitat zu Berlin, Department of Chemistry, Analytical Group
1666 A.Pleshkova, Nesmeyanov Inst.Org.Elem.Cpds, Moscow
1662 Chuck Anderson, Aldrich Chemical Co.
1279 Drug Lab
1105 TNO Volatile Compounds in Food - Chemical Concepts
1093 Organic Synthesis Lab, MSU, Moscow
1045 William W. Christie, Mylnefield Lipid Analysis, Invergowrie, Dundee, Scotland, UK
995 John Halket,Royal Holloway, University of London, UK
898 A. Brossi, LC, NIDDK, NIH, Bethesda, MD 20892
812 Patti Price, Georgia Bureau of Investigation, Decatur, Georgia
797 V.A.KOPTYUG, ATLAS OF MASS SPECTRA OF ORGANIC COMPOUNDS
787 H. Fales, LC, NHLBI, NIH, Bethesda, MD 20892
704 Tox21 Consortium/NIST Mass Spectrometry Data Center, 2012
702 J.E. WILKINSON S-CUBED, SAN DIEGO, CA.
600 Dr. Jiri Zamecnik, DCIEM, North York, Canada, GP Library
566 R.T.HOLMAN,UNIVERSITY OF MINNESOTA
549 Philip Morris R&D
542 AAFS Toxicology Section MS DB Committee, U. Alberta, Canada
522 A.S.Moskovkin,I.M.Karnaukh, Russian Center on Disaster Medicine
495 Finnigan MAT: Toxicological Library (LIBR-TX)
492 B. Derendyaev, Novosibirsk Institute of Organic Chemistry
406 P.A. Leclercq, Lab. Instrum. Anal., Eindhoven Univ. Technol., Netherlands
396 Leung Pu, NIH, Bethesda, Maryland, U.S.
395 A. Lebedev, Chemistry Department, Moscow State University
384 J. Klune, Insect Chem. Ecol. Lab., USDA, Beltsville, MD 20705
354 Insect Chem. Ecol. Lab., USDA, Beltsville, MD 20705
374 G.S. KING, CHEM. PATHOL. DEP., QUEEN CHARLOTTE'S HOSP., LONDON
364 R RYHAGE MS-LAB KAROLINSKA INSTITUTET STOCKHOLM SWEDEN
352 L. Cohen, LC, NIDDK, NIH, Bethesda, MD 20892
343 Darrick Lee, Cayman Chemical, Ann Arbor, MI
333 J.M. Halket, Trace Anal. Unit (TAU), London, UK
332 HD Science, UK
326 N.D.Kagramanov, Moscow, Russia
317 W.Jennings, T.Shibamoto
310 F. TURECEK,HEYROVSKY INST PHYS CHEM & ELECTROCHEM,PRAGUE,CZECH
308 A. Mujsce, AT&T Bell Labs., Murray Hill, N.J., USA
296 O A MAMER, MCGILL UNIVERSITY, MONTRE
295 G. Lange, Inst. Org. Chem., Univ. Wurzburg, Germany
279 TAT
272 K. Kirk, LC, NIDDK, NIH, Bethesda, MD 20892
144 K. Kirk, LBC, NIDDK, NIH, Bethesda, MD 20892
272 Richard Turcotte. Direction des laboratoires d'expertises et d'analyses alimentaires, Quebec, Canada.
262 K. Jacobson, LBC, NIDDK, NIH, Bethesda, MD 20892
259 A.Pleshkova, Moscow,Russia
255 A.Pleshkova, Inst. Org. Elem. Cpds, Moscow
251 Dmitri Zagorevskii, Scientific Instruments & Applications
251 MASS SPECTROMETRY CENTER, UNIV. OF UTAH; ALDRICH
242 J. Daly, M. Garraffo, T. Spande, LBC-NIDDK-NIH, Bethesda, MD
241 S. MARKEY UNIV. OF COLO. MED. CTR., DENVER, COLORADO, USA
235 Dennis Rohrbaugh, CBDCOM/ERDEC, Edgewood, MD
231 H. Luftmann, Org. Chem. Inst., Westfalisch-Wilhelms Univ., Munster, Germany
230 CONTINENTAL OIL CO., PONCA CITY, OKLA, USA
217 N.Klyuev, Institute of Ecology & Evolution, RAS, Moscow
212 S.M. Lee, CA Export Lab. Services, CA, USA
211 J. SHAW, BORRISTON RESEARCH, ROCKVILLE, MD, USA
198 D.G.Hayward MS, Center for Food Safety and Applied Nutrition, FDA, College Park MD.
194 J.VOTH UNIV. OF CALIFORNIA, DAVIS, CALIFORNIA, 95616, USA
192 BAY
189 P. VAINIOTALO, DEPT. CHEM., UNIV. JOENSUU, JOENSUU, FINLAND
188 Susan Richardson
179 ATLAS OF MASS SPECTRA OF ORGANIC COMPOUNDS, V. A. KOPTYUG, ED.
175 V.A. KOPTYUG, ATLAS OF MS OF ORG. CMPDS., ED. 2
173 Dr. P.K. Shah, NYC Police Laboratory, NY
166 W.T.RAINEY OAK RIDGE NATIONAL LABORATORY,OAK RIDGE,TN.USA
162 A.I. Gren, Phys.-Chem. Inst., Acad. Sci. Ukraine, Odessa, Ukraine
162 Dmitri Zagorevskii, University of Missouri-Columbia, Columbia, MO
162 G. Lange, Inst. Org. Chem., Univ. Wurzburg, Wurzburg, Germany
161 A. Brossi, LAC, NIDDK, NIH, Bethesda, MD 20892
160 R.E.Ardrey ET AL.Pharmaceutical Mass Spectra,L.,1985
157 Association of Official Racing Chemists
157 N.W. Davies, Centr. Sci. Lab., Univ. Tasmania, Hobart, Australia
156 Institute of Chemistry, SAS, Bratislava, Slovakia
151 S.A. Rang et al., IR & MS of Unsatd. Hydrocarbons, 1977
149 ChemBridge Corp.
145 MASS SPECTROMETRY CENTER, UNIV. OF UTAH EPA-PTSEL
144 CATALOGUE OF MASS SPECTRA OF PESTICIDES, APRIL, 1975; J. FREUDENTHAL & L. G. GRAMBERG, NAT'L INST. OF PUBLIC HEALTH, THE NETHERLANDS
142 VERIFIN - Methodology
141 Mark Whitten, Florida Museum of Natural History, U. of Florida
139 FOOD RESEARCH REPORT 1XX, K.E. MURRAY & COLLEAGUES, DIV. OF FOOD RESEARCH, CSIRO, AUSTRALIA
137 VERIFINN
135 B.R. PETTIT, QUEEN CHARLOTTE'S MAT. HOSP., LONDON, U.K.
134 J.T. CLERC, ORGANIC CHEM. LAB., ETH, ZURICH, SWITZ.
132 R. P. Adams and V. Dev, Synthesis and GCMS analysis of angelates and tiglates as an aid to identification of these components in essential oils, Flavour and Fragrance Journal, 25, 71, 2010
129 Virginia Division of Forensic Science
127 E. Zissis, LC, NIDDK, NIH, Bethesda, MD 20892
126 Frank Antolasic, RMIT Applied Chemistry Department, Victoria Australia 3001
123 S K.-T. Yu et al. RI and mass spectra of PFP and HFB esters of carboxylic acid, Nat Inst for Petroleum and Energy Research, 1990
122 R. C. DOUGHERTY
119 H. ALDERCREUTZ, UNIVERSITY OF HELSINKI, HELSINKI, FINLAND
117 M.G.HORNING,INST.FOR LIPID RES,BAYLOR COLL.OF MED,HOUSTON,TEX
115 H.LAATSCH,INST. ORG. CHEM.,GEORG-AUGUST-UNIV. GOTTINGEN,W.GER.
111 Pu Leung, NIH, Bethesda, MD
110 D.J.HARVEY UNIV. DEPT. OF PHARMACOLOGY, OXFORD, UK
110 T.A.BLAZER, DU PONT, GIBBSTOWN, N.J., USA
107 OGDEN BioServices Corp., NCI, Rockville, MD 20852
104 USAF ACADEMY, COLORADO SPRINGS, COLORADO 80840; J LLOYD PFLUG
102 J.M. MILLER,BROCK UNIVERSITY,ONTARIO,CANADA
102 L. Tsai, LB, NHLBI, NIH, Bethesda, MD 20892
102 P. BRUCK, MAGYAR TUDOMANYOS AKADEMIA, HUNGARY
101 CSV Ltd., Moscow, Russia
101 FDA, Los Angeles District Laboratory
101 R.W.A.OLIVER UNIV. OF SALFORD, SALFORD, LANCASHIRE, UK
...and thousands of other sources...
MS/MS Library
2023 Details
51,501 Compounds, 60% More than 2020 400 K Precursor Ions – 2.4 M Spectra
Fragmentation Methods: 49,590 HRAM (High Res Accurate Mass) compounds, 50,071 QTOF, HCD, IT-HRAM, QqQ compounds, 49,561 Ion Trap Compounds (Low Res., up to MS4) compounds, 561 APCI HRAM Compounds
Precursor Ion Types: 44,191 Protonated, 19,620 Deprotonated, 14,318 Water/Ammonia Loss, 44,547 Other In-Source Generated
2017 Details
This release contains two MS/MS libraries (nist_msms and nist_msms2), one for small molecules, di and tri peptides and the other for commercially available peptides. It has 652,475 spectra of 123,881 ions of 15,243 different compounds including 1,436 biologically-relevant peptides. See NIST MS/MS for details on the MS/MS library.
Most of the MS/MS spectra have been measured on ion trap and collision cell (qTOF, QQQ, and HCD) instruments using electrospray ionization, although spectra from a number of other instrument types and ionization methods are represented.
The MS/MS Library can optionally be purchased separately at a lower price (with a complete copy of the MS Search software), if you are not interested in the other data in NIST.
Tip:
Even though MS/MS and EI spectra differ and MS/MS spectra are therefore only directly library searched against the MS/MS library, there are still ways to utilize the EI library for MS/MS interpretation. See the article
Structural Identification of Unknowns from Spectra Obtained Using LC/MS and CAD Techniques and EI Mass Spectral Database.
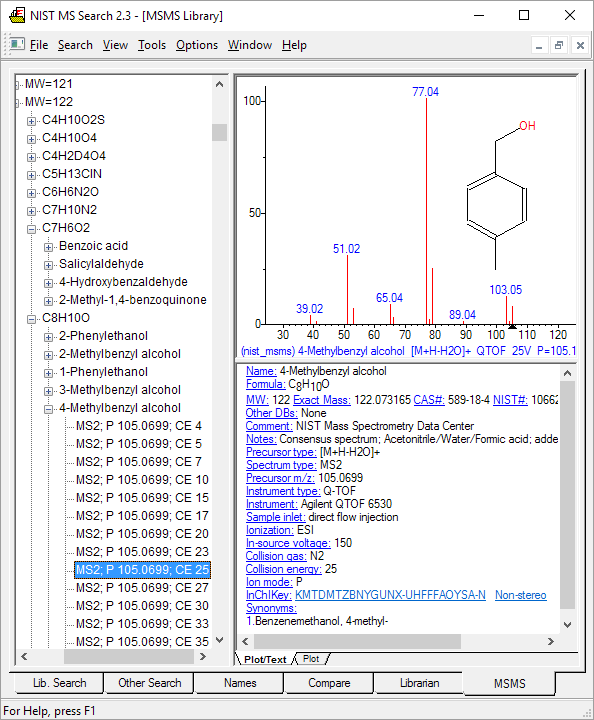
Figure: MS/MS search tab in NIST14. Spectra organized by positive/negative charge, mass, formula, compound name, and acquisition energy/conditions.
Gas Chromatography (GC) Data Library
Gas phase retention data for compounds common to the EI and NIST Retention Data collection are provided with links to the EI library. This involves 492K Kovacs retention indices and corresponding GC methods, column conditions and literature citations for compounds, 153K of which are in the EI spectra library. Data include both non-polar and polar columns. Most values were abstracted from the open literature and then compared to replicate values and estimates to find and remove errors. We also continue to provide estimates of retention indices for most compounds in the library using an extension of a method developed earlier for boiling points [2-3].
GC data includes
- GC column info: column type (Capillary or Packed), column class (Semi-standard non-polar, Standard non-polar, or Standard polar), column diameter (e.g. 0.25 mm), column length (e.g. 30 m), active phase (e.g. DB-5), substrate (e.g. Chromosorb W), phase thickness (e.g. 0.25 um), and carrier gas (e.g. N2/Helium/H2/etc.).
- GC method: description, program type (Ramp, Complex, or Isothermal), Start Temp, End Temp, Start Time, End Time, and Heat Rate
- Retention index data: Retention index type (Normal alkane RI, Linear RI, Kovats RI, Lee RI, Normal alkane RI value specified by scale definition, Lee RI value specified by scale definition) and retention index (e.g. 514).
- Compound identification: Name (e.g. Limonene), Formula (e.g. C10H16), Synonyms (e.g. Cyclohexene, 1-methyl-4-(1-methylethenyl)-), INCHIKEY (e.g. XMGQYMWWDOXHJM-UHFFFAOYSA-N) (lookup), ID (e.g. 33687), CAS Number (e.g. 138-86-3), and NIST NIST Number (e.g. 79639).
Typical GC data for a compound (decanol) is below:
Experimental RI median±deviation (#data) Semi-standard non-polar: 1273±2 (69) Standard non-polar: 1257±3 (73) Polar: 1760±9 (70) Estimated non-polar retention index (n-alkane scale): Value: 1258 iu Confidence interval (Alcohols): 41(50%) 176(95%) iu Retention index. 1. Value: 1256 iu Column Type: Capillary Column Class: Standard non-polar Active Phase: CP-Sil PONA GB Column Length: 100 m Carrier Gas: He Column Diameter: 0.25 mm Phase Thickness: 0.25 um Data Type: Linear RI Program Type: Ramp Start T: 140 C End T: 230 C Heat Rate: 5 K/min Start Time: 10 min End Time: 25 min Source: Cunicao, M.M.; Lopes, A.R.; Cocco, L.C.; Yamamoto, C.I.; Plocharski, R.C.B.; Miguel, M.D.; Junior, A.G.; Auer, C.G.; Miguel, O.G., Phytochemical and antibacterial evaluation of essential oils from Ottonia Martiana Miq. (Piperaceae), J. Braz. Chem. Soc., 18(1), 2007, 184-188. 2. Value: 1255.2 iu Column Type: Capillary Column Class: Standard non-polar Active Phase: DB-1 Column Length: 30 m Carrier Gas: He Column Diameter: 0.25 mm Phase Thickness: 0.25 um Data Type: Linear RI Program Type: Ramp Start T: 40 C End T: 325 C Heat Rate: 3 K/min Source: Sun, G.; Stremple, P., Retention index characterization of flavor, fragrance, and many other compounds on DB-1 and DB-XLB, 2003. ...214 more GC records (omitted from this display)...
The GC data is useful not only for the GC retention index values but also for the GC column conditions and cited literature that goes with it.
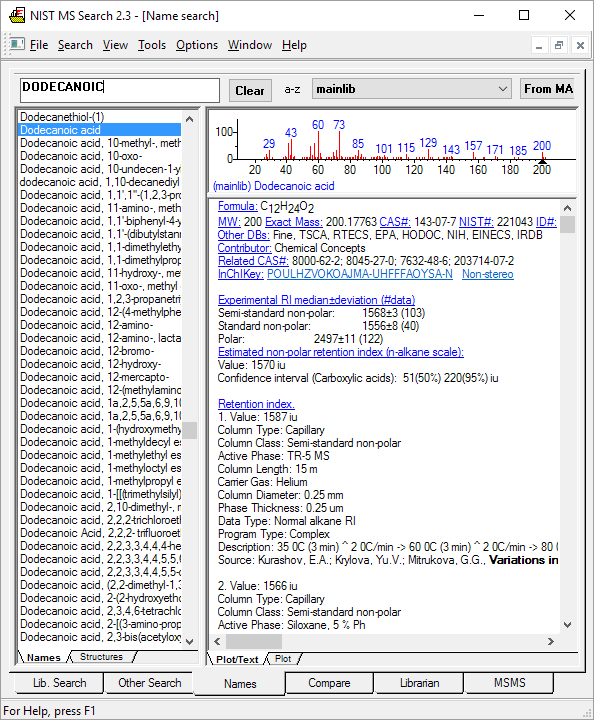
Figure: NIST MS Search software showing GC method and retention index data for a compound with an EI mass spectrum.
See NIST GC/RI for details on this library.
References:
[1]. Babushok, V.I., Linstrom, P.J., Reed, J.J., Zenkevich, I.G., Brown, R.L., Mallard, W.G., Stein, S.E., Development of a database of gas chromatographic retention properties of organic compounds. J.Chromatog. A, 2007, 1157, 414-421.
[2]. Stein, S.E., Brown, R.L. Estimation of normal boiling points from group contributions. J.Chem.Inform.Comput.Sci., 1994, 34, 581-587.
[3]. Stein, S.E., Babushok, V.I., Brown, R.L., Linstrom, P.J., Estimation of Kovats retention indices using group contributions. J.Chem.Inf.Modeling, 2007, 47, 975-980.
NIST MS Search Software
(Note: See What's New in NIST23 for more recent changes.)

Figure: NIST MS Search software
The updated, full-featured NIST MS Search Program for Windows has a full range of integrated tools. The new updated version of this widely used, full-featured software is designed for identifying compounds from their mass spectra and for exploring mass spectral libraries. It also contains tools for deconvoluting gas/liquid chromatograms and interpreting mass spectra.
- Library Searching via Multiple Search Modes
- Chemical Identity
- Structure Similarity
- Impurity Reduction
- Neutral Loss
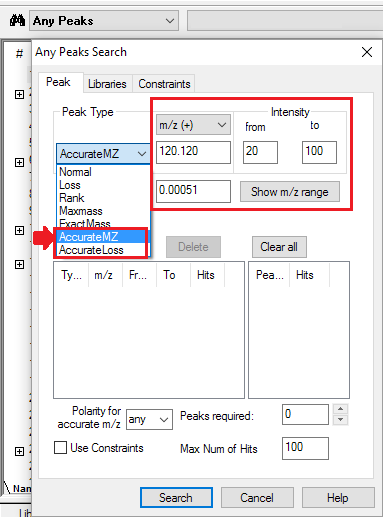
- Four Peak Types
- Library Building
- GC/MS Deconvolution
- MS Interpretation
- Chemical Substructure Analysis
- Chemical Substructure Identification
- Molecular Weight Estimation
- Chlorine/Bromine Analysis
- Isotope/Formula Generator
- Integration with multiple Instrument Data Systems
Overview
This software provides a flexible means of accessing data in the NIST and User libraries including:
- chemical identification by optimized, documented spectrum-matching methods,
- analyzing mass spectra of compounds not in the library using library data,
- finding spectra with pre-selected characteristics,
- viewing spectra by name, formula, CAS registry number, molecular weight, or ID number,
- User library building and maintenance, and
- integration with a number of commercial GC/MS data systems and spectral analysis tools.
When the program is first started, seven tiled windows appear on the screen (the Desktop), each with its own data and behavior. The behavior of any Window may be modified by making it active (clicking on it) and then pressing the AdjustWin button at the bottom of the Desktop. As you become familiar with program operation, you may wish to change the dimensions of some Windows or even close some of them to create a custom Desktop. When the program is restarted, it will begin with the most recent Desktop. To save a window-arrangement Desktop for future use, select Desktop from the Menu Bar and then Save As from the resulting Menu. Prior Desktops may be restored using one of the predefined names or your own name. Such arrangements describe the geometry and type of information shown in each Window, not the actual data contained in it. To restore a previous hit list, select it from the list at the top of the Hit List Window.
Library Searching - Identify unknown compounds and substructures using fully documented and optimized procedures, or search by a wide range of compound and spectral properties.
Library Building - Maintain your own libraries, add your own chemical structures and search using the same optimized procedures developed for NIST.
Flexible User Interface - Set multiple Desktop configurations with up to seven independently configured windows to examine search results and match your needs.
Use with Your Instrument Data Systems - Direct transfer between a number of commercial data systems and the NIST Search Program.
Adding User-Drawn Structures
Users may import their own chemical structures
with selected user spectra. This is done in the Tools/Librarian section
of the program by connecting a user-drawn structure in standard MOL-file
format with a user spectrum. Such structure-drawing programs are
widely available (for example, ISIS/Draw may be freely downloaded
from http://www.mdli.com/prod/freesw.html). As before, if a user
spectrum is given its CAS registry number and the Main Library has a structure
for it, this structure will automatically be shown with user spectra unless
the user has attached an imported structure to the spectrum.
Aids for Automation and Reporting
A variety of methods for automated searching and reporting of results
are available. From the File menu selection, if Print automation
is on, printing will follow each library search. A set of print options
is also available from the "User Search Options" Window (select Search,
then User spectrum). This is of particular use when using this NIST
Program with other data acquisition programs.
What's New in NIST23
Main scope of changes:
- The NIST 23 MS/MS Tandem library has grown to ~50,000 compounds (2.4M spectra) - all at multiple fragmentation energies and conditions.
- NIST 23 EI library (GC/GS) has added spectra of 40,000 compounds with their retention indices to their EI Library, most of them selected by, measured and evaluated at NIST.
- NIST has made significant enhancements to their metadata and their software, Mass Spectral Search 3.0.
New features of the program and database are:
- Major Spectrum Type choice (EI, Tandem, Peptide, All) helps to suppress display of search parameters most probably not used.
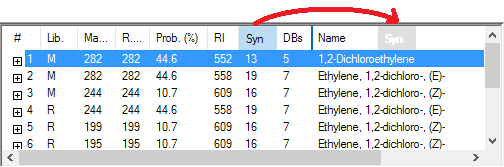
- Name search sorting has been increased from 16 to 249 characters; the list of names has been split into two panes: compound list in the upper pane and spectrum list in the lower pane. All spectra derived from a selected compound are shown in the spectrum list (EI: replicate, derivatives, stereoisomers; Tandem: energy, instrument, fragmentation type).
- A list of 'Other Databases' (compound references) for each compound has been significantly extended. See file Other_DBs.xlsx
- Added a new search method - Partial Spectrum Search - for searching with abbreviated spectra.
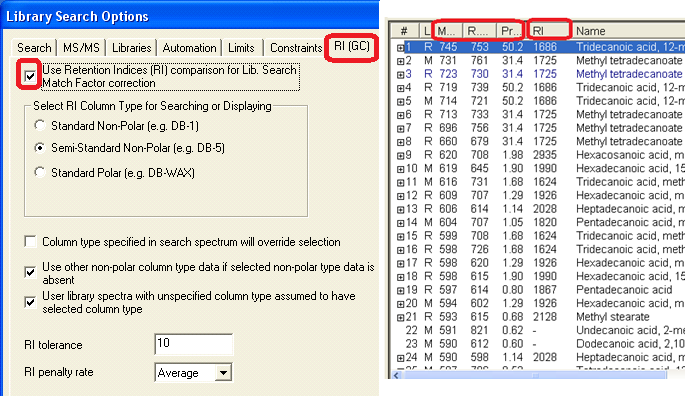
- All retention indices are displayed in the EI hit list. They are used for EI search with RI match factor correction. If experimental data is not available for a compound, a reliable AI predicted value is used in its place.
- Hovering a mouse pointer around a peak in a spectrum plot displays peak annotation.
Changes to the program (compared to MS Search v2.4 / NIST 20):
- Term "In-source/EI with accurate ion m/z" replaced with "HiRes No Precursor"
- "Penalize rare compounds" EI library search option has been retired.
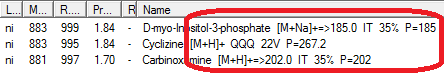
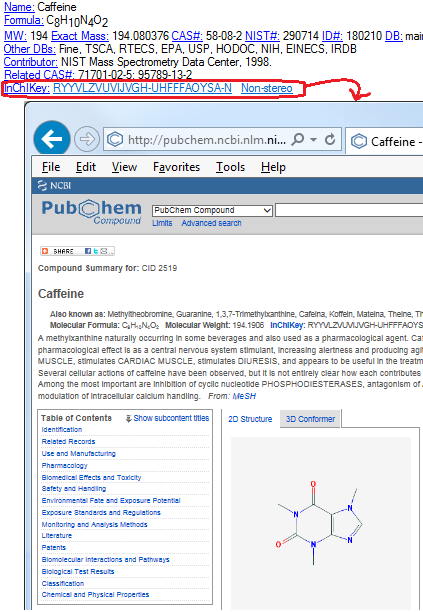
- PubChem InChiKey search has been replaced with Google search.
- Major mass spectral text file extension has been changed from .MSP to .MSPEC. Extension .MSP is still recognized.
Features added to recent versions (NIST 20 MS Search v.2.4 and earlier):
- Major expansion of coverage of analytically important compounds of all classes, based on documented importance of each compound.
- For the first time ever, almost all spectra measured and fully evaluated entirely at NIST.
- Modernized and enhanced evaluation procedure allowing for multiple evaluations of spectra.
- Enhanced comparison of replicate spectra and 'best of replicates' selection. The entire library is now based on chemical structures using the NIST-developed InChIKey representation.
- Enhanced quality control using greatly improved MS Interpreter program that links ions to originating structures in the precursor molecule.
- Enhanced method for assisting the examination of results having many spectra generated by Tandem spectra. See "What's New in MS Search Program" topic in MS Search help for more details.
Some noteworthy features of the program and database in recent versions:
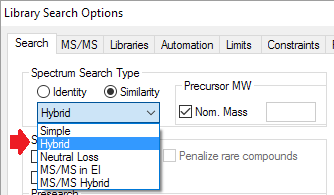
- A new search type, the 'Hybrid Search' for all spectrum types, enable finding many more similar compounds in the hit list. It requires a precursor mass.
- A new neutral loss search is available that eliminates restrictions on the maximum loss considered.
- Improved algorithms for scoring high mass accuracy searches that account for sparse spectra.
- Peaks shifted in the 'Hybrid Search' are shown before and after shifting.
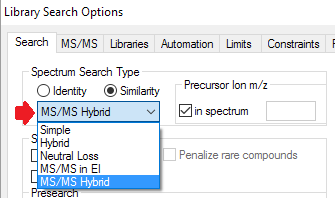
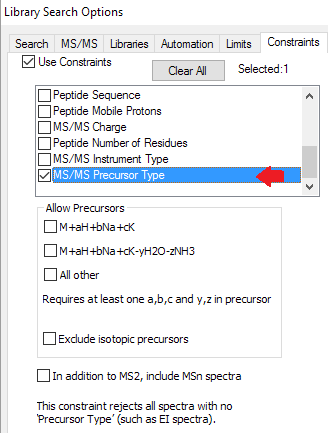
- New filters for tandem mass spectra to limit hits by charge, precursor type, instrument, msn, and mass accuracy.
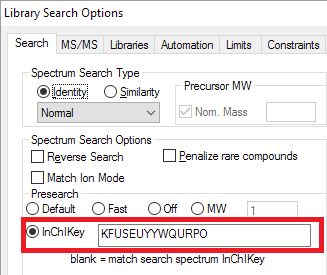
- An 'InChI' presearch that limits identification to compounds with the same structure as the search spectrum.
- Searching by exact precursor m/z.
- MS/MS browsing window and MS/MS library
- New type of mass spectrum search: MS/MS Identity search for searching for MS/MS spectra in MS/MS libraries. The updated Lib2NIST converter is a tool for creating MS/MS libraries. To create a MS/MS library, run Lib2NIST and check "MS/MS spectra only" option. For this search to work, the MS/MS spectra must contain precursor m/z values.
- Derivatives for GC/MS can be represented as their precursor molecules using a large number of derivative types.
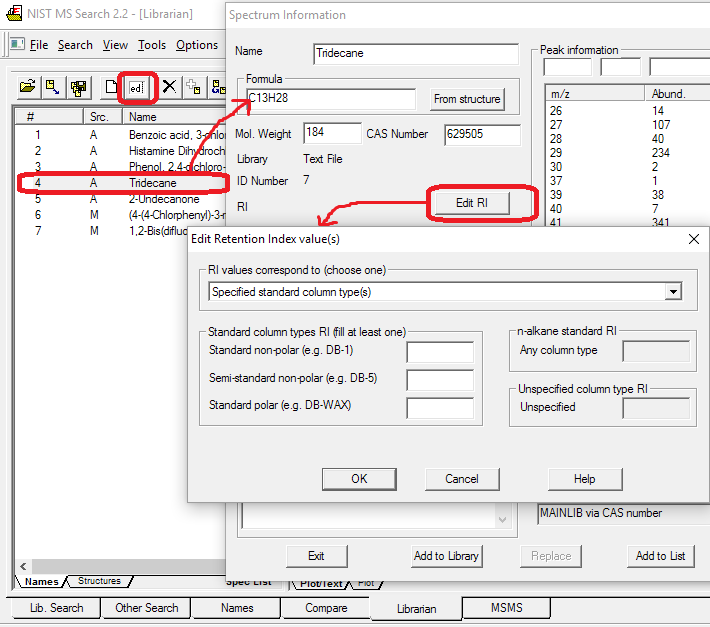
- Retention indexes in some of Main and Replicates libraries spectra. Automatic Retention Index estimation from chemical structure (right-click spectrum text window, select Properties, Retention Indices). Polar columns were new to the 2008 release.
- Structural replicates display (in addition to usual CAS number matching) in Main and Replicate libraries, including a new feature: displaying derivatives and a wider range of tautomers. Select matching method in Options, Replicates.
- Structure searching - the program will find all chemical structures similar to the structure of the search spectrum. The searching can be done using structural data from the NIST MS database, from the user database or from a structure drawn in most chemical drawing packages. A structure may be exported to chemical drawing software. The Structure similarity search algorithm has been changed: now it includes homologue recognition that generates a match factor of 1000 for homologs. You will need to rebuild structure search databases in user libraries (as you usually need to do after editing a user library containing structures.)
- Searching in the Contributor/Comment spectral records and extracting tagged fields into the displayed spectrum text information.
- Drag and Drop - Data can be dragged and dropped between windows. Point mouse to the name of the compound in the spectrum plot window, press and hold the left mouse button, drag the spectrum and drop it into Spec List or Compare Window.
- User Library Structures - This feature has been present for some time, but many are unaware of it.
- Compare Views - There are a number of different ways for you to visually compare two spectra.
- Direct interaction with a number of chemical drawing packages - the clipboard can be used with some of them, most can be used by use of intermediate files.
- The NIST MS Search program takes advantages of many normal Windows conventions. Look at the context sensitive options on the right mouse button click and on buttons at the top of the window.
- Thermodynamics-based MS Interpreter program version 3.0. This program now works for both low and high mass accuracy spectra and permits using a mouse to select parts of a chemical structure for mass and formula determination and analysis.
New in NIST 20
New in NIST 17
New in NIST 14
New in NIST 11
New in NIST 08
New in NIST 05
New in NIST 02 and previous versions
Automated Mass Spectrometry Deconvolution and Identification System (AMDIS)
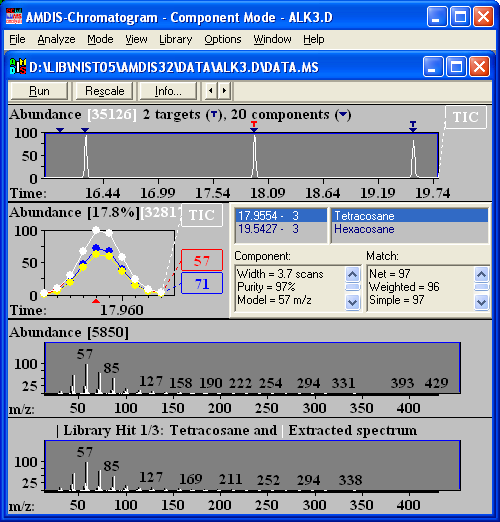
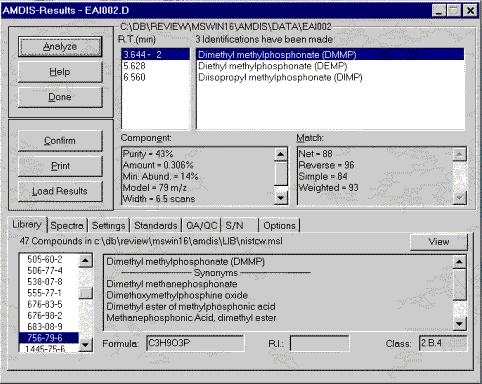
The AMDIS software extracts pure component spectra from complex GC/MS or LC/MS data files and searches against specialized libraries or the NIST library. This module was developed at the National Institute of Standards and Technology (NIST) for the critical task of verifying a major international treaty, the Chemical Weapons Convention. After two years of development and extensive testing it is now being made available to the general analytical chemistry community. AMDIS can operate as a "black box" chemical identifier, displaying all identifications that meet a user-selectable degree of confidence. Identification can be aided by internal standards and retention times. Libraries can be built directly from analyzed data files or from spectra in the NIST/EPS/NIH Database. AMDIS can also serve as a preprocessing tool for the GC/MS data files, automatically performing noise reduction for all components. It permits traditional library searching for any selected component. A flexible interface is provided to assist the analysis of complex matrices.
Included as a separate utility, AMDIS attempts to reconstruct original mass spectra for individual components in arbitrarily complex GC/MS and LC/MS reconstructed total ion current (RTIC) chromatograms; and, if a target library is provided, can directly identify target compounds. AMDIS is especially useful when an RTIC chromatographic peak represents multiple components. Regardless of each component's concentration, pure mass spectra are deconvoluted for analyses. AMDIS was developed by NIST for the Defense Weapons Agency (Department of Defense) for verifying compliance with a major international treaty (Chemical Weapons Convention) ratified by the United States Senate in 1997. In order to meet the rigorous requirements for this purpose, AMDIS was tested against more than 30,000 GC/MS data files accumulated by the EPA Contract Laboratory Program without a single false-positive for the target set of known chemical warfare agents. While this level of reliability may not be required for all laboratories, this shows the degree to which the algorithms have been tested.
AMDIS has been designed to reconstruct "pure component" spectra from complex RTIC chromatograms even when components are present at trace levels. For this purpose, observed chromatographic behavior is used along with a range of noise-reduction methods. AMDIS is distributed with specialized libraries (environmental, flavor and fragrance, and drugs and toxins), that were derived from the NIST Library. AMDIS has a range of other features, including the ability to search the entire NIST Library with any of the spectra extracted from the original data file. It can also employ retention index windows when identifying target compounds and can make use of internal and external standards maintained in separate libraries. A history list of selected performance standards is also maintained.
As of version 2.73, AMDIS reads data files in the following formats:
- Agilent ChemStation (*.D)
- Agilent MS Engine (*.MS)
- Bruker (.MSF)
- Finnigan GCQ (*.MS)
- Finnigan INCON (*.MI)
- Finnigan ITDS (*.DAT)
- HP ChemStation (*.D)
- HP MS Engine (*.MS)
- INFICON GCMS (*.acq)
- JEOL/Shrader (*.lrp)
- Kratos Mach3 (*.run)
- MassLynx NT (*.*)
- Micromass (*.)
- mzXML/mzData (*.*) (new in NIST11)
- NetCDF (*.CDF)
- PE Turbo Mass (*.raw)
- Shimadzu MS (*.R##)
- Shrader/GCMate (*.lrp)
- Varian MS (*.MS)
- Varian SMS (*.sms)
- Varian XMS (*.xms)
- Xcalibur Raw (*.raw) - requires Xcalibur be installed
AMDIS may also be downloaded separately.
Additional NIST MS Tools: Mass Spec Interpreter
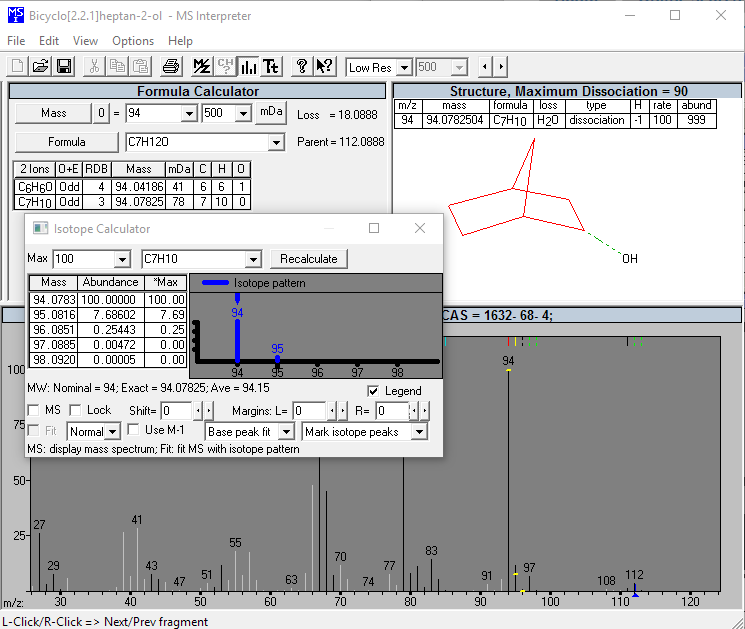
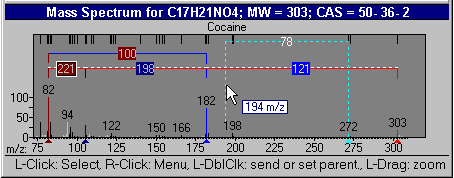
Figure: NIST MS interpreter
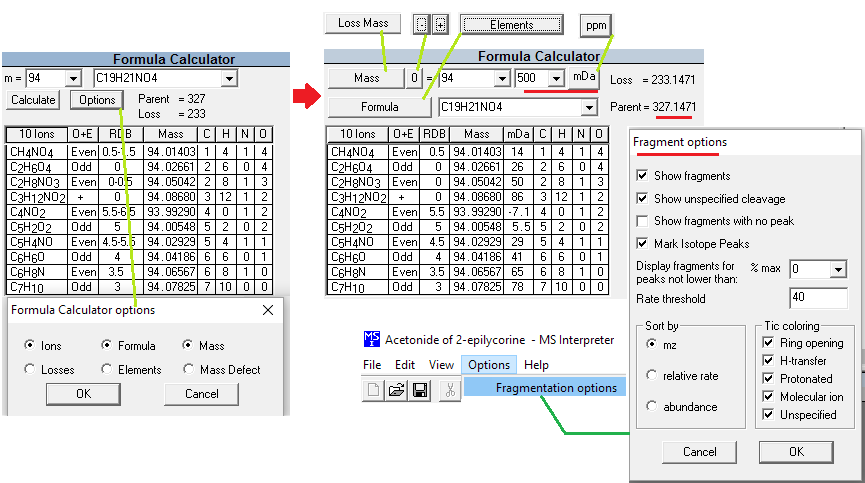
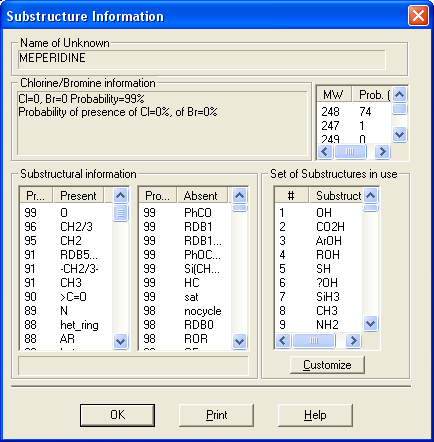
This utility enables the user to examine neutral losses, isotope patterns and possible chemical formulas along with computer assisted chemical structure/spectral analysis. Starting with Version 1.5, the program has offered a unique means of interpreting spectra of compounds not identified in the Library by the User spectrum search. This is most useful when no acceptable matching spectra are found in a User spectrum search of the NIST Library. At this point, by selecting Substructure Information from the Tools menu, the current hit list is analyzed and statistical information on the composition of the unknown is derived from the hit list. For instance, the probability that any of a range of substructures are present or absent are listed (phenyl, acid, ester group, double bond, heteroatom, etc.) along with an estimate of the molecular weight and chlorine/bromine content (the latter is based only on the spectrum itself).
This tool was developed to aid NIST evaluators in their analysis of mass spectra. In one integrated program, it permits a wide range of calculations on a mass spectrum using--if available--a proposed chemical structure. Spectra and structures are associated in the library facility of the Windows Search Program discussed above, and the program is activated from the Tools menu. Peaks in the spectrum originating as a logical fragment of the molecule are marked, and corresponding fragments may be highlighted. It also allows the analyst to keep track of important neutral losses, both from the parent or a derived ion, and to readily compute possible formulas for any peak or neutral loss and isotopic patterns as desired. Observed isotopic clusters can be compared to theoretical predictions subject to a number of user-specified constraints.
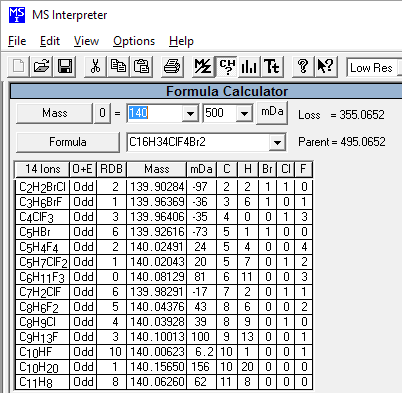
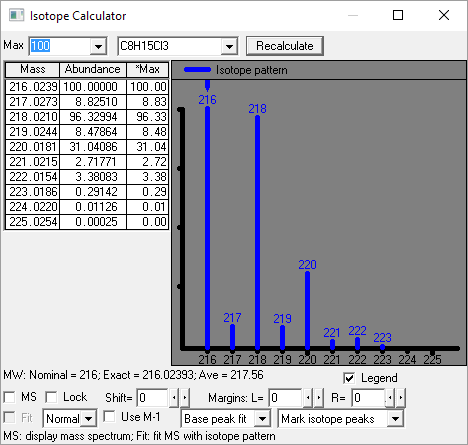
Figure: NIST Formula and Isotopic Pattern Generator
System Requirements
Operating system requirements (NIST 23):
- Windows 11, 10, 8.1, 8, 7, Vista, XP SP3 (both 32 and 64-bit editions) are fully supported.
- Windows 2000 is not compatible with NIST 17 MS Search (NIST 14 MS Search does work). ChemStation .L format will work.
- Windows NT 4 is partly compatible with NIST14/11
- Windows NT 3.x, Me, 98, 95, 3.x are not compatible with NIST14/11 MS Search. ChemStation .L format might work.
Complete installation (libraries, NIST MS Search program, and AMDIS) with both EI and MS/MS libraries requires about 9.37 GB of free hard disk space, but selecting EI only will save considerable disk space.
Mass Spec Data System Compatibility
Instructions by MS manufacturer:
- Agilent ChemStation (or MassHunter) - The Agilent version of NIST is recommended for GC/MS ChemStation or MassHunter, as this provides the most seamless integration with Agilent data systems. This version includes a copy of the NIST library formatted in the Agilent library format (*.L) with structures, for searching by Agilent's PBM library search routines. Also, Agilent ChemStation (*.D) and MS Engine (*.MS) can be imported into AMDIS using the standard version of NIST. For LC/MS ChemStations (which don't use the .L library format), the standard version of NIST will work just as well.
- Thermo (Finnigan) - The standard version of NIST can be used with Xcalibur and Chromeleon, which integrates directly with the Xcalibur data system. Also, Xcalibur Raw (*.raw), Finnigan GCQ (*.MS), INCOS (*.MI), ITDS (*.DAT) can be imported into AMDIS using the standard version of NIST. The NIST library in the Thermo Spectral ID is available in the multiformat version of NIST.
- Varian Saturn - The standard version of NIST can be used. Also, Varian (*.MS), SMS (*.sms), and XMS (*.xms) can be imported into AMDIS using the standard version of NIST.
- Waters MassLynx - The standard version of NIST is recommended, at least for MassLynx versions > 4.0. On search, the spectra is automatically transferred into the NIST MS Search window for library searching. (Older MassLynx versions only support the native MassLynx library format, which is available in the multiformat version of NIST and Agilent/Other version. In fact, newer MassLynx versions continue to support this method as well, but the information displayed has been reported to be more limited than with NIST MS Search--e.g. in terms of information displayed, search time, and 8-digit CAS number limit.) MassLynx NT (*.) can also be imported into AMDIS using the standard version of NIST.
- Waters Empower - Contact us for details. Please tell us what version of Waters Empower you are using.
- Shimadzu: NIST in the native Shimadzu GCMSSolution library format is available in the multiformat version of NIST, allowing searching (.qgd) directly from the Shimadzu software. The native Shimadzu library format has only been tested on recent 2.x versions of GCMSSolution software. Chemical structure diagrams are not included in the native Shimadzu format itself, though you can still browse structures in the NIST MS Search software. Alternately, using only the standard (or any other) version of NIST, it is possible to import old Shimadzu QP5000/QP5050 "CLASS-5000" software MS (*.R*) files into NIST AMDIS, for searching instead from the NIST MS Search software, but this approach does not work with the newer .qgd files without first converting the .qgd to .cdf (e.g. using MASSTransit).
- PE TurboMass (*.raw) files can be imported into NIST MS Search via AMDIS using the standard version of NIST. NIST MS Search may also be directly integrated with the PE TurboMass software with the standard version (but call us for instructions). Also, NIST in TurboMass library format (.IDI/.IDB/.PDB/.PDI), where TurboMass will do the library searching without MS Search, is included the multiformat version of NIST.
- Bruker .MSF files can be imported into AMDIS using the standard version of NIST.
- INFICON GCMS (*.acq) can be imported into AMDIS using the standard version of NIST. PC software for Hapsite® supports NIST and AMDIS.
- JEOL/Shrader/GCMate (*.lrp) can be imported into AMDIS using the standard version of NIST. Shrader also has direct interfaces into the standard version of NIST. Contact us for details.
- Kratos Mach3 (*.run) can be imported into AMDIS using the standard version of NIST.
- Micromass (*.) can be imported into AMDIS using the standard version of NIST.
- Others/NetCDF (*.cdf) can be imported into AMDIS using the standard version of NIST. NetCDF is a vendor-neutral format supported by a number of data systems and GC/MS format conversion software (e.g. GC/MS File Translator and MASSTransit).
(The above list is somewhat of a moving target. Please drop us an e-mail if anything here is incomplete.)
Reference
- NIST User Manual
- Further information is in the NIST Help file (Help menu).
Additional Links
- NIST Instruction Videos, by James Little - NEW VIDEOS!
-
Mass Spectral Library Gets Modern Upgrade to Deal With Drug Deluge - Forensic Maganize interviews Steve Stein on NIST 17:
- - - "In [this release], we have something called the hybrid search, which is a new kind of mass spectrometry search method that can reliably identify compounds—even if they are not in your library—that differ from your library compound by one chemical group that doesn’t affect the fragmentation, like fluorine or methyl," explained Stephen Stein, the NIST chemist who oversees the library. "Many of the designer drugs have derivatives. With one [compound] in the library, you can theoretically identify hundreds of others. If the functional group doesn’t change, the signature doesn’t change, so you can match that. It increases the coverage of your library with just an algorithm. - - - "One interesting thing about a big library is all the compounds that could masquerade as false positives are probably in a big library. So, you can show the uniqueness of your spectrum only by searching against a large library," he said. But, he’s quick to point out libraries don’t identify compounds—people do." - - -
- "NIST 11: What's New and What Value Does it Offer?"
by O. David Sparkman, Separation Science, sepscience.com, 2011.
- Part 1 - The history and current status of NIST 11 and its value to analytical scientists.
- Part 2 - The history and current status of NIST 11 and its value to analytical scientists (more).
- Part 3 - In Part 3, the use of the searches in the Other Search tab view is examined. These searches can be beneficial to the identity of compounds from mass spectra obtained by ionization techniques other than EI and from data that provides a higher accuracy of the measure m/z value than is usually available for EI data.
- Part 4 - This installment in the series on NIST 11 is about the Incremental Name Search, replicate spectra, and the NIST GC Methods, and Retention Index Database.
- Separation Science MS Solutions also has additional AMDIS related articles
- A "Little" Mass Spec and Sailing blog - various resources and tips on NIST by James Little / Eastman Chemical Co, e.g.: NIST searching at Eastman
- MS Calc Pro - simple molecular structure drawing, fragmentation, MS digitization, and isotope calculation tool, with integration with NIST MS Search
- software: Tal-Aviv Molecule Identifier (TAMI) - The TAMI software automatically analyzes your measured mass spectra, based on the analysis of the measured isotopic pattern of the molecular ion (Isotope Abundance Analysis – IAA), and links with NIST's MS Library to confirm or reject its identification results. This significantly enhancing the confidence level in the NIST library identification. In addition, TAMI independently provides the sample elemental formula in case it is not in the library.
- MolView - free web-based ".MOL" file (molecular structure) editor. ".MOL" files can be imported/exported from NIST MS Search.
- MS courses
- Old:
Demo Version
There are two ways to evaluate NIST:
Download NIST 17 Demo. The demo differs from the full version of NIST mainly in that it contains a very small subset of the EI database and a somewhat older version of the software (about 2300 spectra). Also, this is a demo does not contain the Agilent integration (ChemStation/MassHunter macros nor NIST in the ChemStation .L format), although you can load Agilent .D files into the included NIST AMDIS program, which in turn integrates with NIST MS Search. The demo also does not contain MS/MS spectra, although there is a separate peptide demo available for that (NIST MS/MS peptide demo). Use our online search to check whether compounds are present in the full database.
Note: For some MS data systems that search for the NIST MS Search program location in the "c:\windows\win.ini" (or "c:\winnt\win.ini") file, you might need to edit that file in a text editor to change the line "[NISTMSDEMO]" to "[NISTMS]" like the real version of NIST.
Download AMDIS 2.71 (July 2012) (free) (AMDIS is included in NIST 17 demo and need not be downloaded separately)
Other files:
- 2011-12-28 - An updated ChemStation macro (NISTSrch.mac) is available to avoid interference in situations where both ChemStation and another search system (e.g. Xcalibur or Masslynx) are used simultaneously on the same computer (by reusing the autoimp.msd file). [download ZIP] (contains NISTSrch.mac and install instructions).
Ordering
NIST23 now available!
| Part No. | Description | Price EA |
Order |
|---|---|---|---|
| NIST 23 - New Licenses | |||
| NIST23 | NIST 23 Standard Version | $2856.60 | |
| NIST23AG | NIST 23 Agilent Format Version. (Includes Standard Version) | $3180.60 | |
| NIST23MF | NIST 23 Shimadzu/Multiformat Version | $6154.00 | |
| NIST 23 - Upgrades | |||
| NIST23U | NIST 23 Standard Version, UPGRADE | $1647.00 | |
| NIST23AGU | NIST 23 Agilent Format Version, UPGRADE. (Includes Standard version) | $1897.56 | |
| NIST23MFU | NIST 23 Shimadzu/Multiformat Version, UPGRADE | $4138.00 | |
| * Download only. | |||
|
|
|||
Which version to buy? Call or e-mail us if you have any questions about which version to buy. In summary:
- The standard version of NIST (NIST23) is recommended for most users (except for Agilent ChemStation/MassHunter and Shimadzu GC/MS Solutions). It includes the NIST library formatted in NIST format, which is the native format understood by the NIST MS Search program, which integrates with a number of data systems (e.g. Varian Saturn, Thermo Xcalibur, Waters MassLynx version >= 4.0). GC/MS, LC/MS, and MS spectra from many data systems can also be imported indirectly into AMDIS (see above) and then library searched via the AMDIS-NIST MS Search program integration.
- The Agilent format version of NIST (NIST23AG) is recommended primarily for most Agilent ChemStation/MassHunter users. It includes everything in the standard version plus the NIST EI library formatted in the Agilent .L library format for library searching directly from Agilent software (e.g. Data Analysis PBM search).
- The Multiformat version of NIST (NIST23MF) is recommended for Shimadzu GCMSsolution users in particular. It includes everything in the standard and Agilent versions of NIST plus the following additional native formats: Shimadzu GCMSsolution, Waters MassLynx, and Thermo Spectral ID.
- Other formats may be available upon request. Further details are in the System Requirements tab above.
Other databases available: The NIST mass spectral database is a strongly validated and extensively used MS library of general compounds recommended for most all users. Users looking for a larger library or libraries dedicated to certain classes of specialty compounds may wish to augment their NIST database with one of the Wiley MS libraries.
(Previous names: NIST 2020/NIST20/NIST 2017/NIST17/NIST 2014/NIST14/NIST 2011/NIST11/NIST 2008/NIST08)
(Older part numbers: NIST 17: NIST20/NIST20AG/NIST20MF/NIST20U/NIST20AGU/NIST20MFU/941010/941010HP/941010MF/941010UG/941010HPUG/941010MFUG, NIST 14: 841010/841010HP/841010MF/841010UG/841010HPUG/841010MFUG)
 Search Names in Database Check whether a certain compound or class of compound exists within the NIST 23 database.
Search Names in Database Check whether a certain compound or class of compound exists within the NIST 23 database. Article By O. David Sparkman Evaluated and Expanded for Quality Figure 1. The NIST Mass Spectral Search Program with all seven of its Windows displayed. Added Features for Quality Prior to 1998, it had been six years since NIST released its last version of the NIST/EPA/NIH Mass Spectral Library. During that period, NIST has completed a ten-year project to completely evaluate the Library. As this process progressed, NIST was able to generate a number of spectra and acquire several important collections o...
Article By O. David Sparkman Evaluated and Expanded for Quality Figure 1. The NIST Mass Spectral Search Program with all seven of its Windows displayed. Added Features for Quality Prior to 1998, it had been six years since NIST released its last version of the NIST/EPA/NIH Mass Spectral Library. During that period, NIST has completed a ten-year project to completely evaluate the Library. As this process progressed, NIST was able to generate a number of spectra and acquire several important collections o...Brochure
 Update Registration NIST current version: The current version of NIST MS is 2020. Notifications on future NIST versions: New versions of the NIST MS database come out roughly every three years (e.g. 1998, 2002, 2005, 2008, 2011, 2014, 2017, 2020), with upgrades available for purchase. If you wish to be notified upon availability of new versions of the NIST database and upgrade offers, please fill out the form below. Details will also be announced on our NIST web page, at the ASMS meeting, and in our general mail l...
Update Registration NIST current version: The current version of NIST MS is 2020. Notifications on future NIST versions: New versions of the NIST MS database come out roughly every three years (e.g. 1998, 2002, 2005, 2008, 2011, 2014, 2017, 2020), with upgrades available for purchase. If you wish to be notified upon availability of new versions of the NIST database and upgrade offers, please fill out the form below. Details will also be announced on our NIST web page, at the ASMS meeting, and in our general mail l...A few features of NIST08 MS Search that you might not know about.